
Python as a calculator
Apart from executing single commands in the command-line, more advanced actions can be taken through the execution of a script. A script, also known as the source code, represents a series of commands, which have the aim for the computer to perform a task. Computer programs are built with such scripts and this process is called computer programming. Scripts are written in a human-readable text format. In the field of computer programming, many different programming languages exist for writing source code.
Two general groups can be distinguished when classifying programming languages: compiled and interpreted.
Compiled programming languages are translated into another programming language by a compiler. Through this process an executable file can be compiled with instructions for the computer. Run times of programs written in compiled programming languages are smaller. However, the development cycle of these languages is slower, because everything from the script needs to be compiled before it can be executed. Examples of such languages are: C, C++, Java, Fortran, Go, etc.
With interpreted languages instructions are executed directly by an interpreter in a step-by-step format and are not first translated into another programming language. Programs based on source code that is written with an interpreted language run slower, but the development cycle of such a script is more flexible. Interpreted languages consist of Perl, Python and R.
In bioinformatics four main programming languages are used to write scripts:
Python can be started directly from the command-line by typing python. Version 2.7.6. is installed on the bmw.gbiomed.kuleuven.be server. Python takes your commands line by line. To exit Python type exit().
>>> points to the interactive mode of Python

Python as a calculator
Data files can also be used in Python on the command-line, just simply transfer your data files to the server (via Bitvise for Windows or Cyberduck for Mac OS).
A script is written as a text file with a text editor. If the script is written in the Python programming language, it is executed by Python, which acts as an interpreter. Script written for python have the extension .py.
The first line of a script always starts with #!, which is called a Shebang, followed by a specification for the programming language you are using. Here, we will use:
#! usr/bin/ev pythonFollowed by this comes the programming code. ___
Hints and tricks:
This exercise is replicated from the book: Practical computing for biologists. For more information on Python programming, check: Haddock SHD, Dunn CS. Practical computing for biologists. Sunderland (Massachusetts U.S.A.): Sinauer Associates, Inc.; 2011.
dnacalc.py will represent a program to calculate the melting temperature Tm of a certain DNA sequence.
Step 1: define a DNA sequence to check the Tm for (less then 14 nucleotides)
Step 2: Calculate the nucleotide counts
Step 3: Calculate Tm according to the following formula

Tm calculation

Python DNA calc
Step 4: How many nucleotides are in your sequence?

Python DNA calc
In this example the DNA sequence is considered a variable. In Python variables store a value and are case-sentive and changeable. The assigning of a value to a python variables occurs through specifying the variable by typing "=".
Up to now, in this script our variables are: DNASeq and SeqLength.
Different types of variables can be distinguished: string / integer / float /list / dictionary.
String: text
Integer: natural numbers
Float: real numbers
List: list of items
Dictionary: list of items ("keys") with an assigned value
Set: unique list
Boolean: True/False
Objects
Python is dynamically typed, so it infers the type of variable for the user. Opposed to this is a statically typed variable, whereby the type needs to be defined by the user.
The three main variable types:

Python variable type
For DNASeq the variable type is a string.
For SeqLength the variable type is an integer.
Every variable type has different operators/functions available:
integer: 3 * 3 = 9
integer and float: 3 * 3.0 = 9.0
string and integer: "a" * 3 = "aaa"
string: len("aaa") = 3
When handling these variable types, errors can occur. Therefore, caution is necessary.
float: 7.0 / 2 = 3.5
integer: 7 / 2 = 3
integer (float): float(7) / 2 = 3.5
string with an integer: "7" / 2 = TypeError
string with an integer: len(7) = TypeError
To determine the type of a variable i:
>>> i = 200
>>> type(i)
<type 'int'>
>>> type (i) is int
TrueHints and tricks:
 ___
___
Step 5: How many of each nucleotide is present in your DNA sequence? Use count() as a function to determine this.
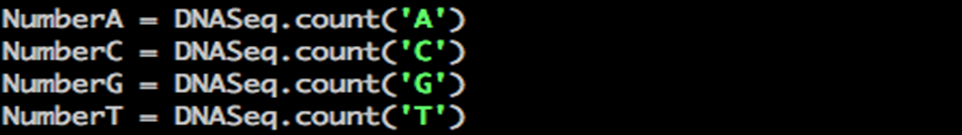
Python dnacalc nucleotide count
Difference len() and count()
To determine the total sequence length earlier, the function len() was used (step 4). In step 5, the function count() is used in combination with a variable, which is called a method. What is the difference between a function and a method in Python?
Function are used for a broad application. Methods are object oriented. Here, DNASeq.count() is an object oriented method for the DNA sequence.
 ___
___
Hints and tricks:
.format().format() is defined by arguments, which represent the variables you want to output{0}: represents argument number 1
{1}: represents argument number 2
..., etc.
.format(argument1, argument2 )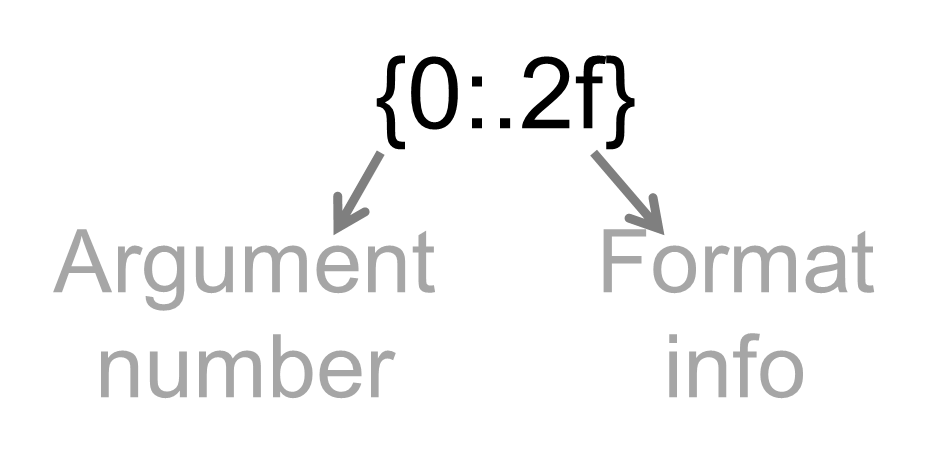

 ___
___
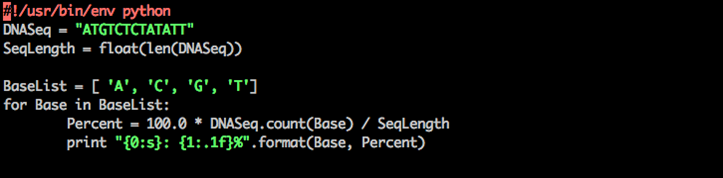
Step 6: print the percentage of A, C, G and T nucleotides in your sequence
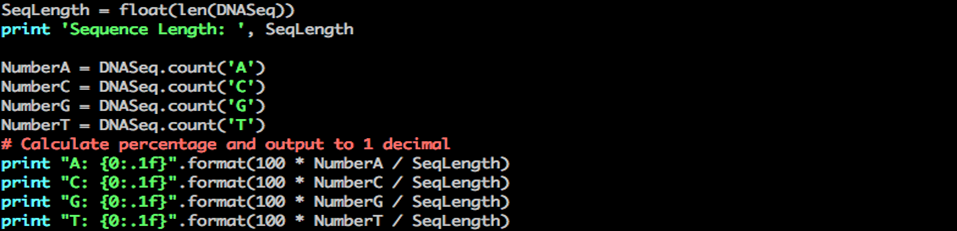
Python percent nucleotides
Step 7: Get input from the user

Python raw input
raw_input() = returns the input provided by the user as a string
Check and correct the input via following operations on a string:
upper(): Make sure all nucleotides are upper case
replace(): Replace something by something else
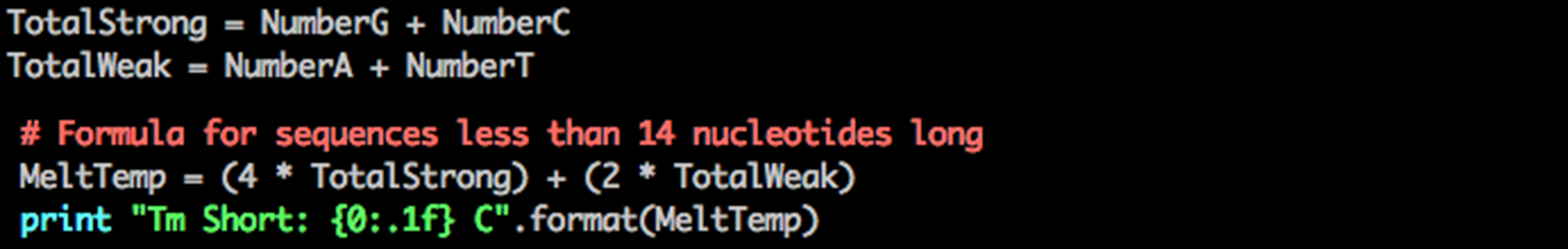
Python Tm short sequence
You now have a script to calculate the melting temperature for DNA sequences with less than 14 nucleotides. But what if the DNA sequence has more than 13 nucleotides?
In this case, another formula is used. Let your script make a decision:
Use the if/elif/else construct.
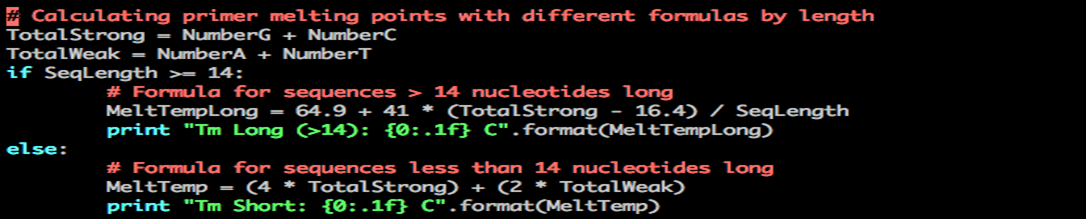
Python Tm long sequence
Hints and tricks:
 ___
___
To make a decision, you also need to specify which situations need to be distinguished from eachother: if (sequence >= 14) or else (sequence < 14).
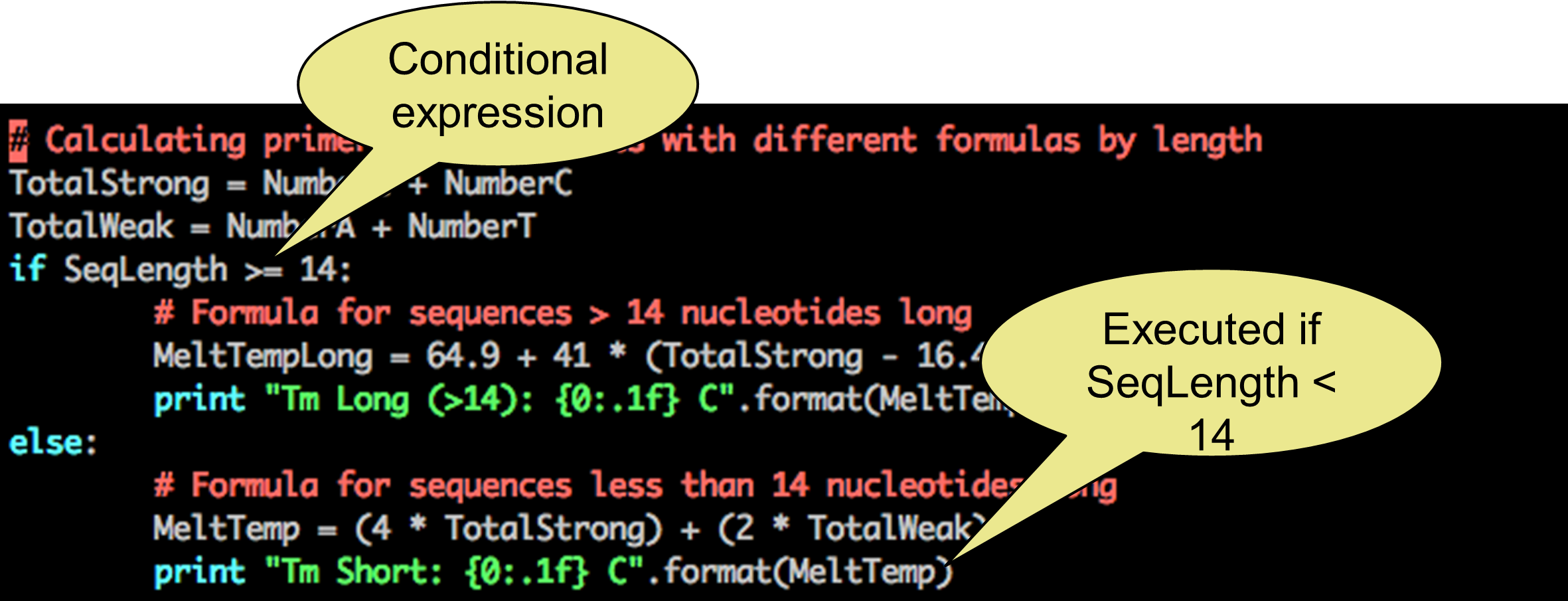
Python conditional expressions
This is done by adding conditional expressions (be aware of variable types when using conditional expressions):

Python conditional expressions
Final step: You have created a Python script, which allows you to enter a DNA sequence and the program calculates the correct melting temperature. The final output represents for example the total sequence length or the Tm of the total sequence. Intermediate outputs include the percentage of each nucleotide. Tm is different for long ( number of nucleotides >=14) or short ( number of nucleotides < 14) sequences and the script identifies the correct formula based on the length of the sequence.

Python dnacalc total script
Hints and tricks:
Copy this script to your own working directory to compare (use . for your current working directory)
cp /tmp/python/dnacalc.py .In order to debug your script: check for capitalization, read the error, read the script, which part of the code is responsible for the error and try to understand what went wrong. Use Google to search for solutions.
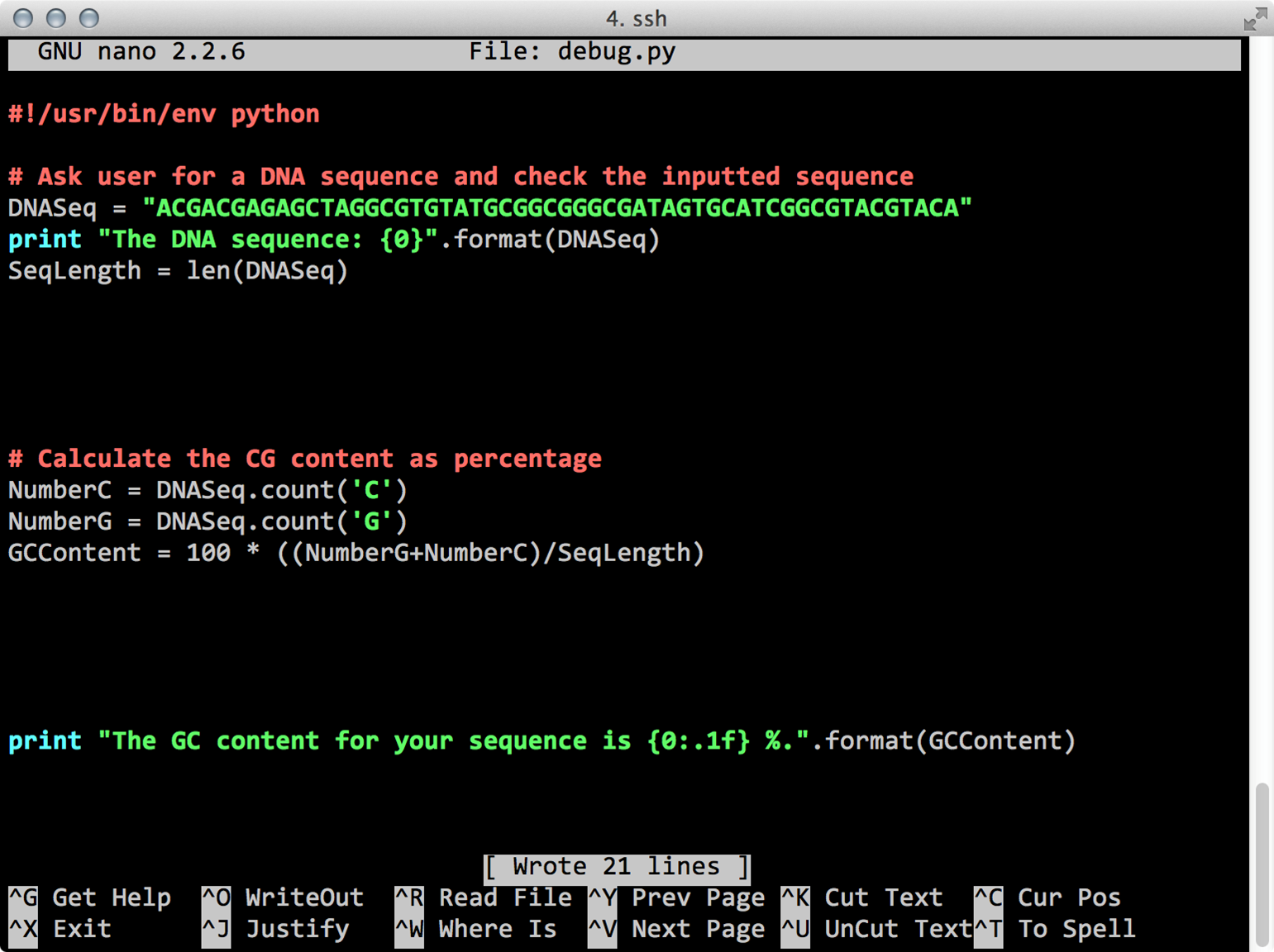
Python debug
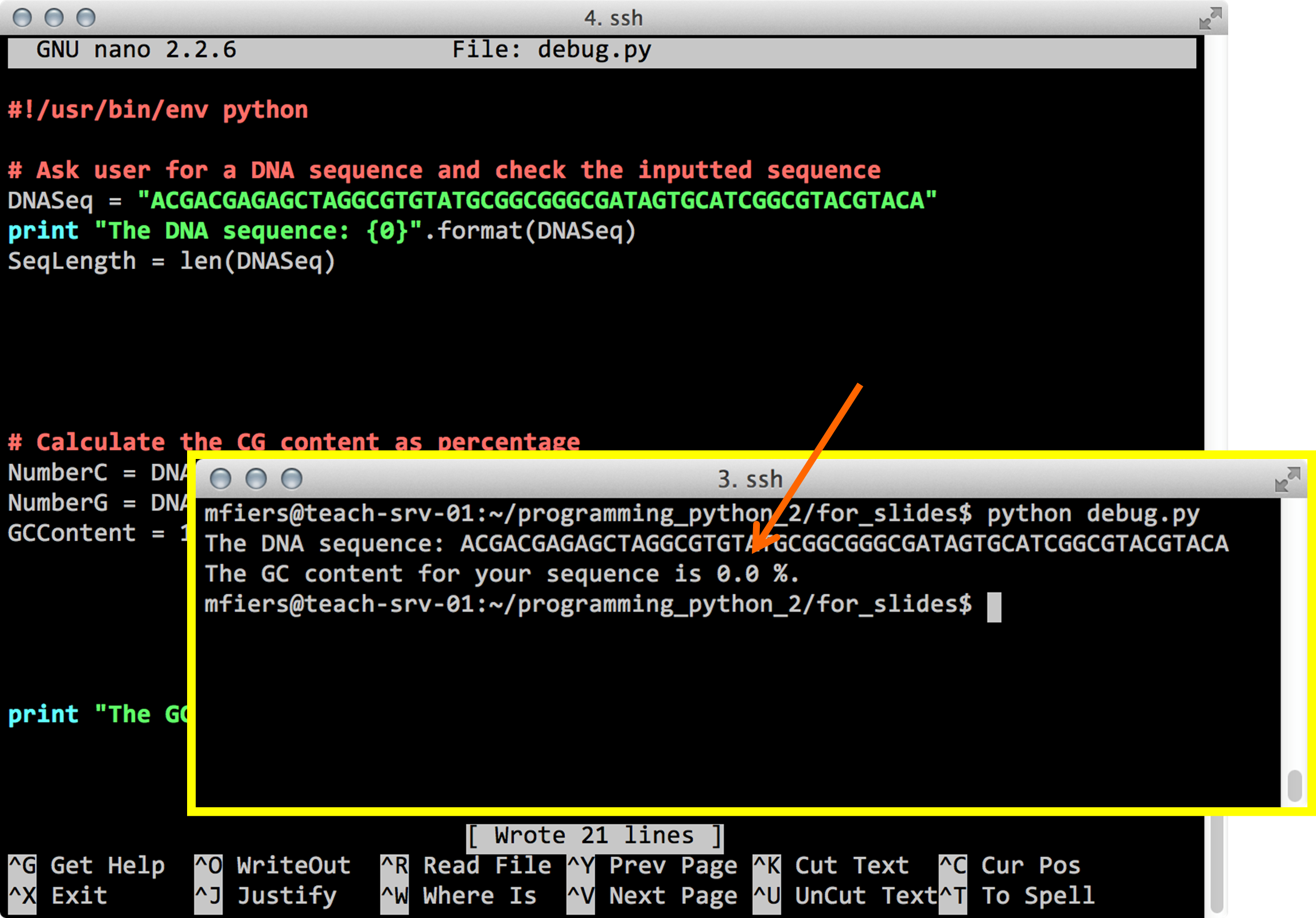
Python debug
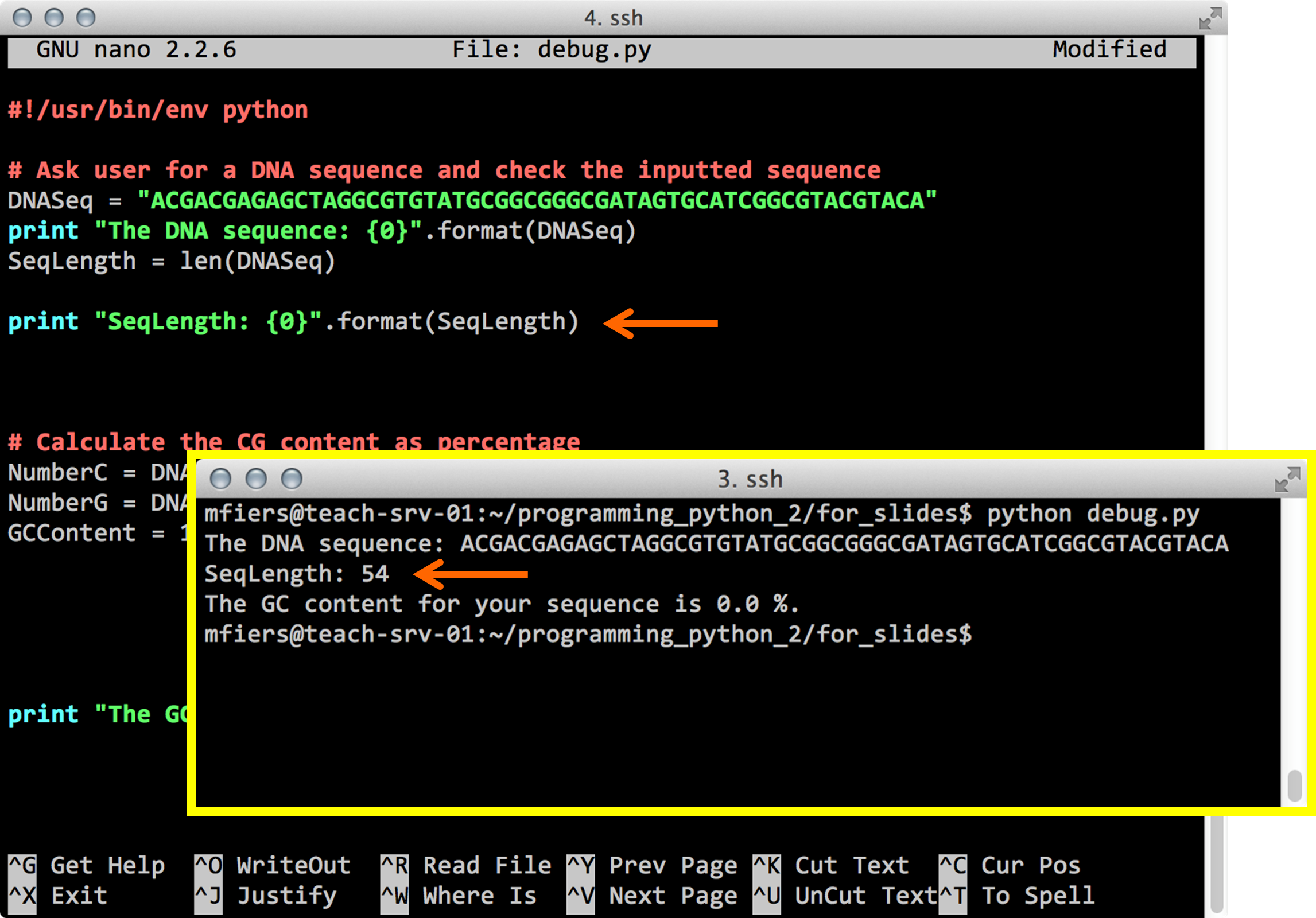
Python debug
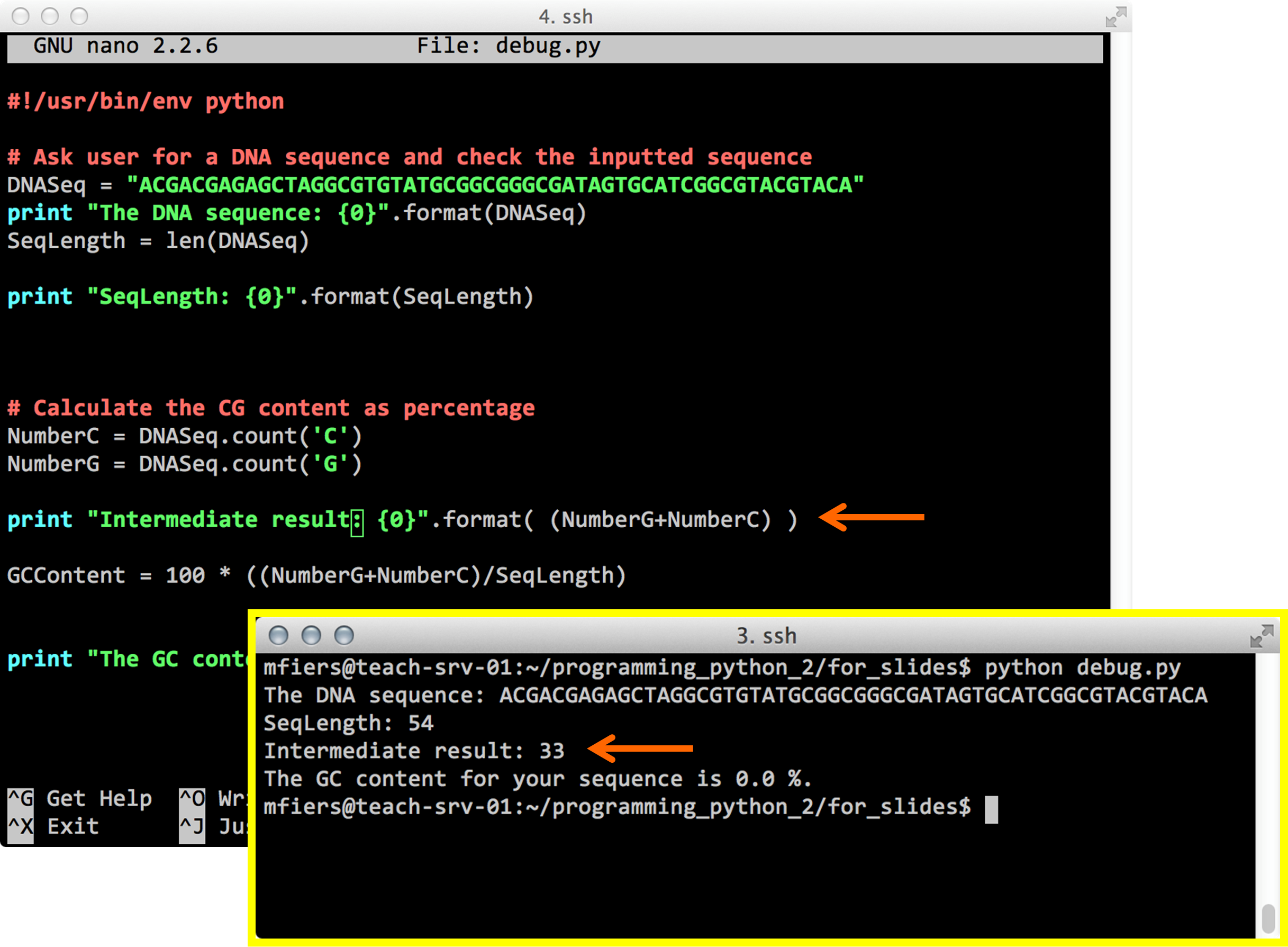
Python debug
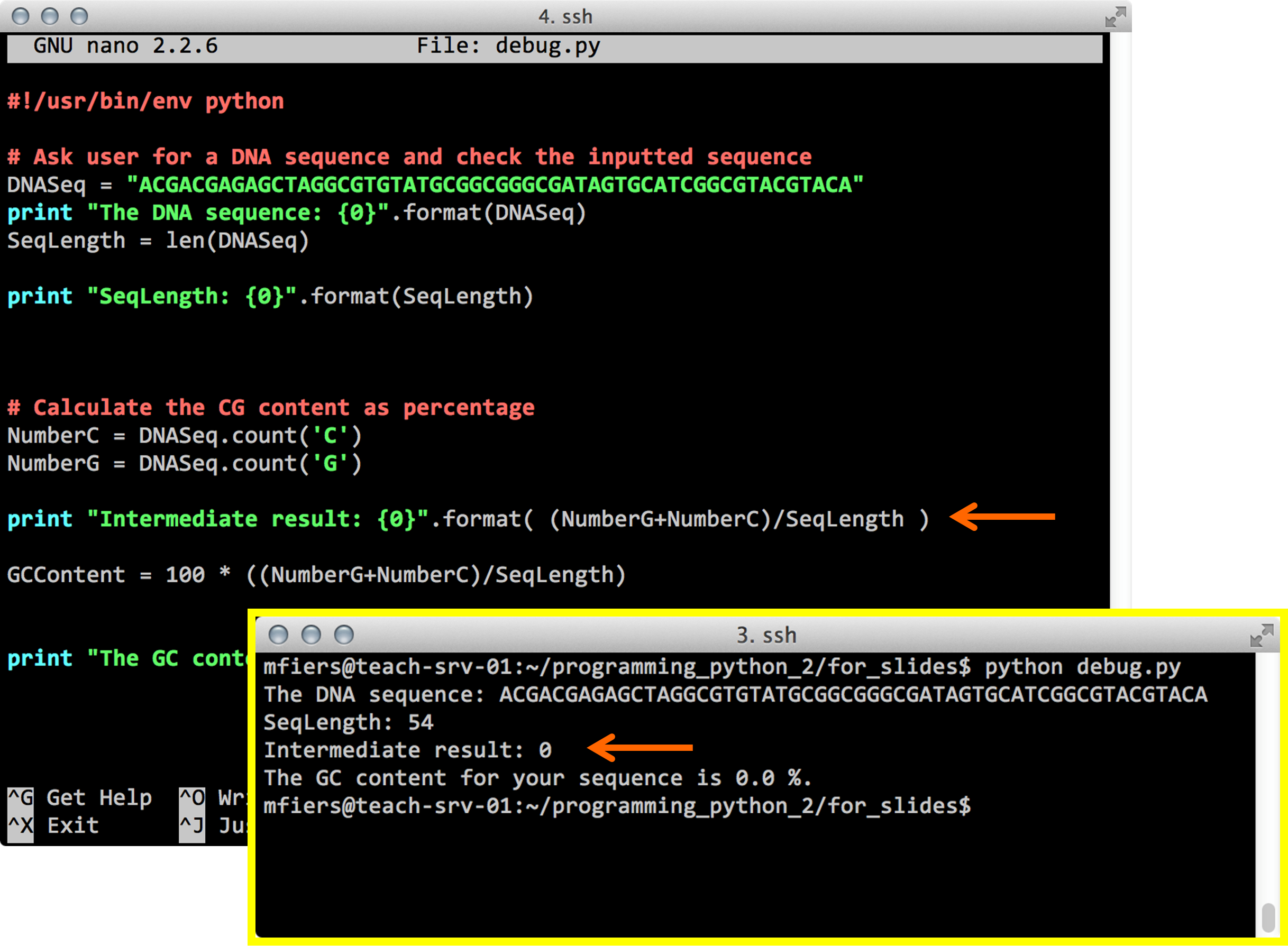
Python debug
Hint:
Think about variable types!
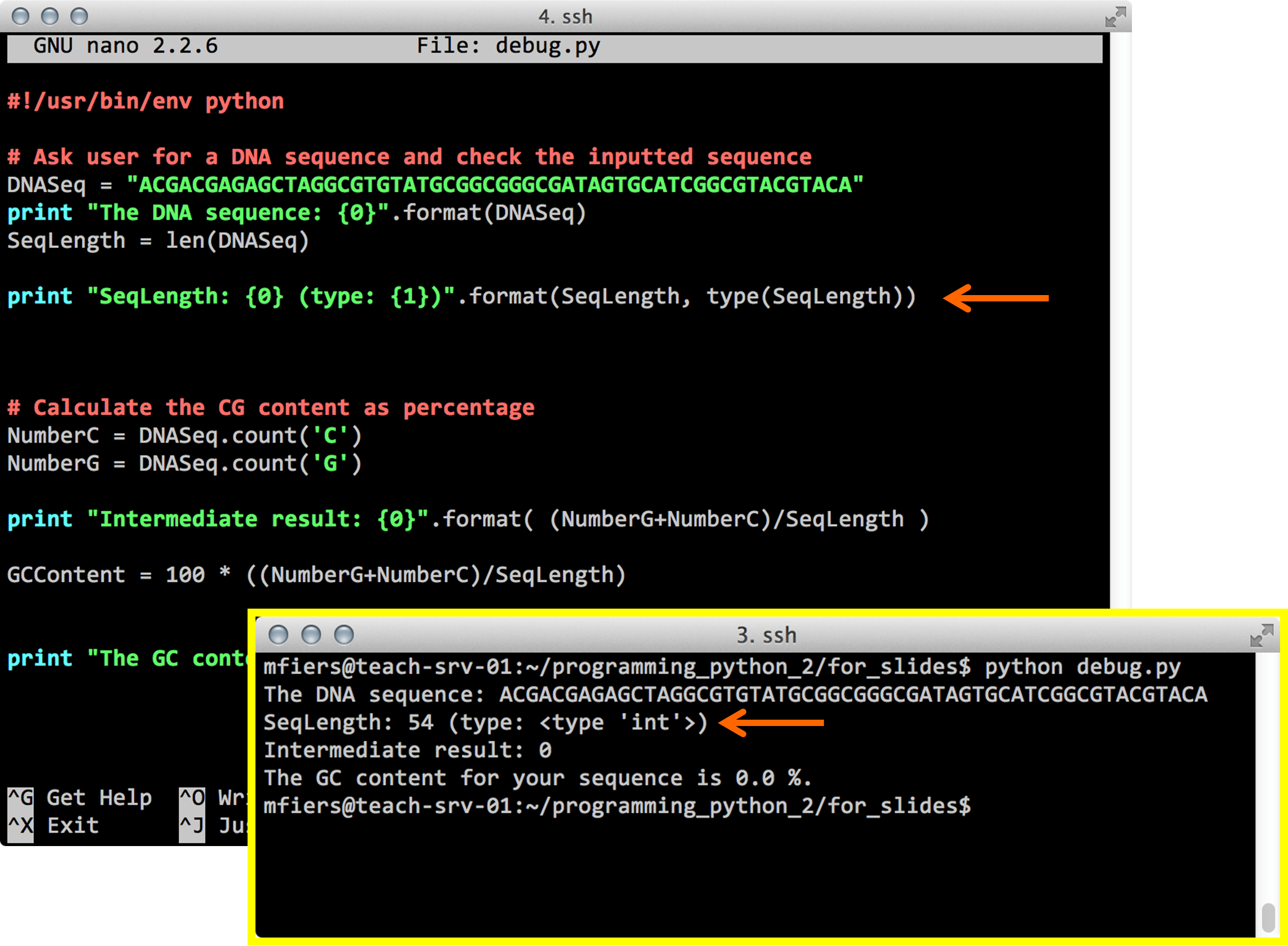
Python debug
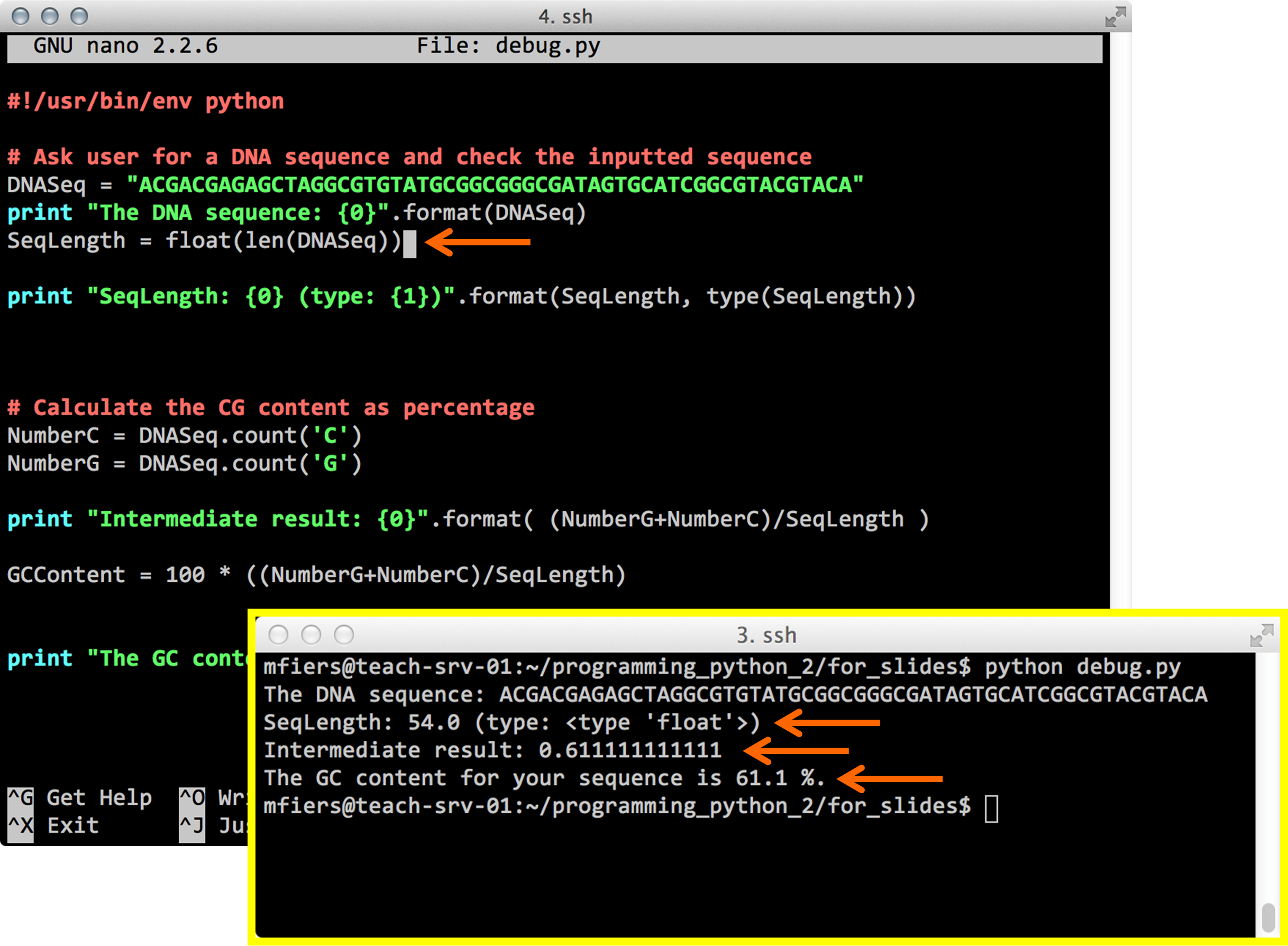
Python debug
As a variable in Python, you can also define a list of values.
list = [value0, value1, value2, ..., value x].append(value3, value4, ..., value y): appends value(s) to the existing list
.pop(value0): removes the value(s) and returns the value(s) to the user
As a final step, you can reverse the list and print the elements:

Python list
Make a list of numbers from 0 to 12 (12 not included in steps of 2):

Python list slices
For each element in the range from 0 to 13 (13 not included in steps of 3), print the element:
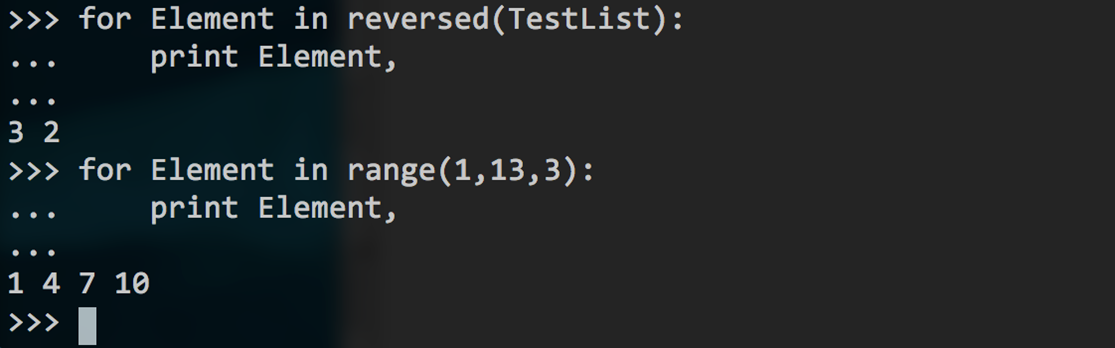
Python loops
Python loops
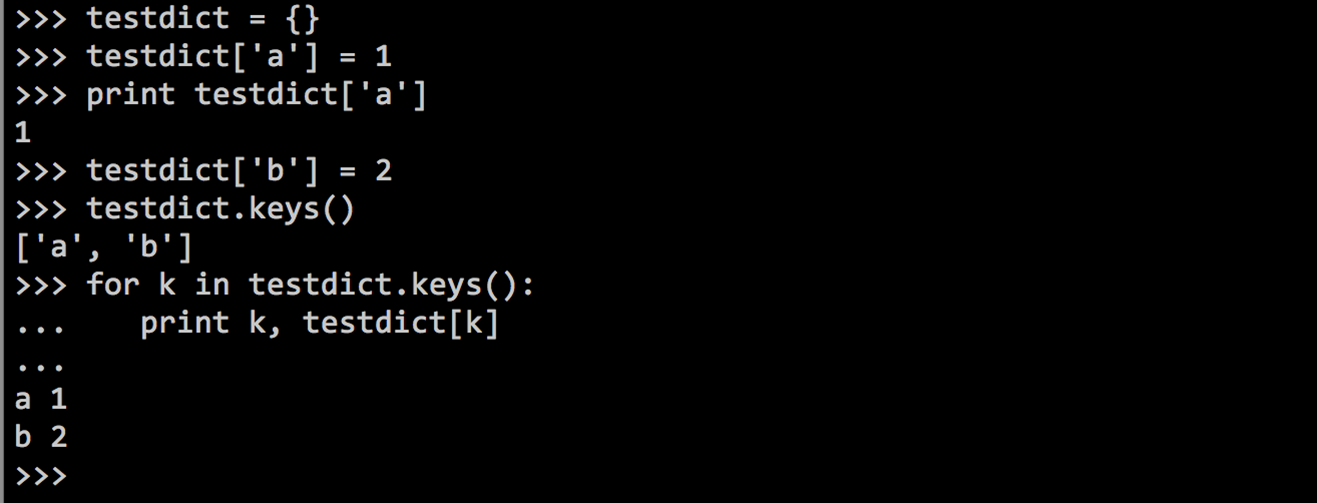
A list can be used to assign values to each element ("key") in a list.

Python dictionaries

Python dictionaries
Python dictionaries
With dictionaries you can make for example a codon table for amino acids:
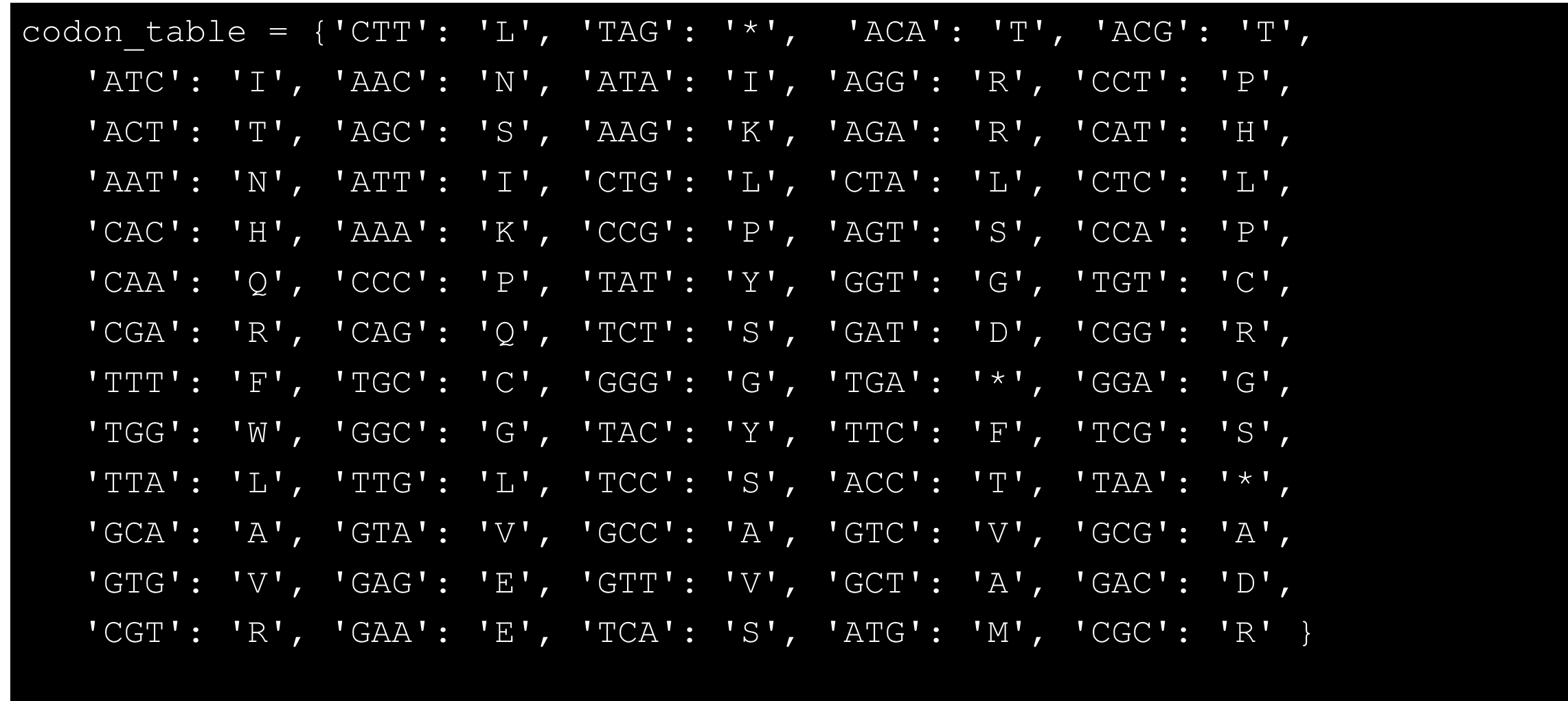
Python codon table
Python is an object-oriented programming language. Objects can be a variable and a function at once. Variables and functions are linked to the objects via classes.

Python reading data

Python reading data

Python writing data
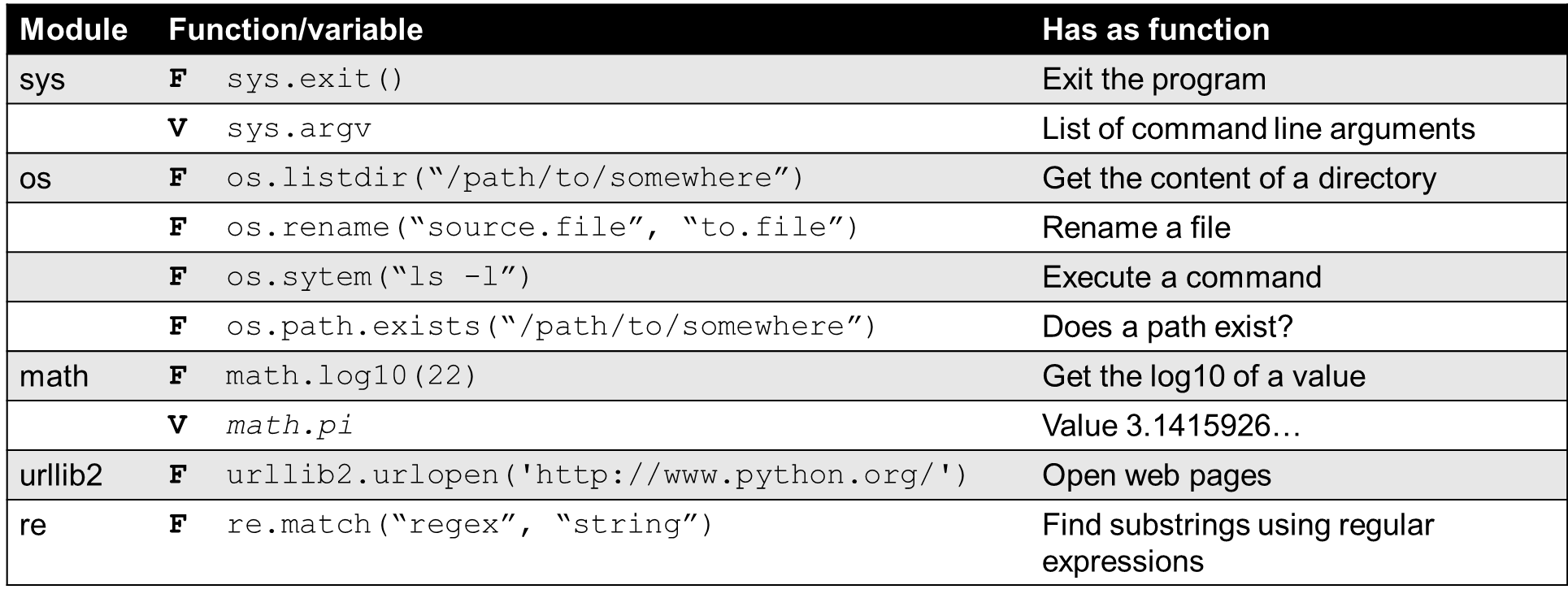
Modules in Python are files with a .py extension, which implement a set of functions. With the use of modules more functionality in your program can be implemented. An overview of multiple Python modules:
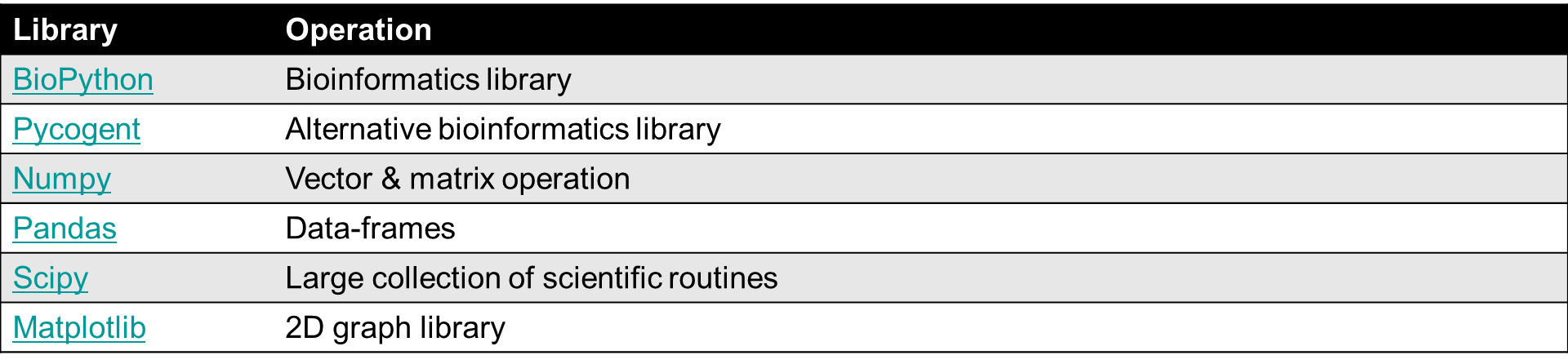
Python modules
Many more are available online: https://pypi.python.org/pypi.
Python itself provides some built-in modules. A summary can be found here: http://docs.python.org/2/library.
Python built in modules
In order to use these modules, you will need to import them with the import statement in Python. When using modules in your program/script, you will need to put the import statements of the modules at the top of your script.
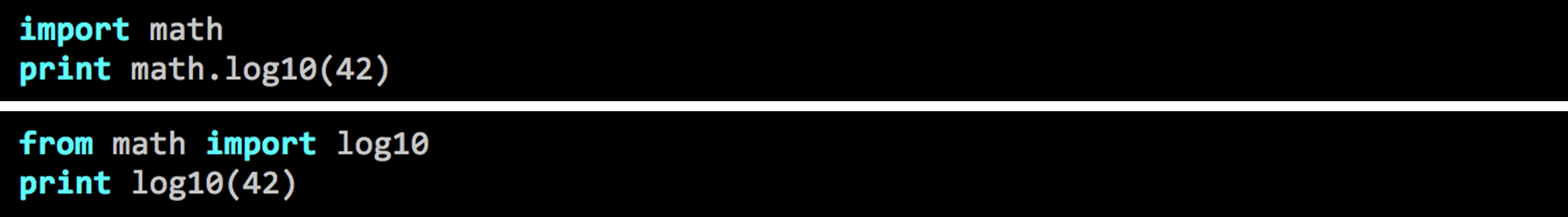
Python import modules
Step 1: read a FASTA file with a DNA sequence from disk
Step 2: convert it to a protein sequence

CDS to protein
Step 3: write FASTA file of protein sequence to disk
example sequence:
/tmp/python/braf.fastaIPython offers an interactive shell for running Python code. Jupyter offers for example a notebook function for your Python code, but also for other programming languages. This allows you to run Python code through a webbrowser.
http://ipython.org/ https://jupyter.org/
IPython/Jupyter
Biopython is collection of modules and tools that are available for Python programming. These tools are specific for the computational biology or bioinformatics field with tools for sequence alignment, sequence motifs, sequence annotation, reverse complement of a sequence, etc. Biopython is compatible with a wide variaty of data types from bioinformatics. Biopython offers a web interface or can be used via the command-line, e.g. BLAST can be run locally or online.
For more information: http://biopython.org/wiki/Main_Page.
Biopython has its own datatypes, called objects. For example:

Biopython seqrecord
As is mentioned in 'Chapter 6: Alignments' alignments can reveal similarity between two sequences and between this can imply common ancestry and function. Alignments are often performed in the field of bioinformatics. Different types of alignments, algorithms and software is available.
Exercise 3.6. BioPython: GC content, NCBI download and alignment of two sequences