
Bitvise SSH Client Login
To analyze data, either generated by your own experiments or retrieved from biological databases, multiple software tools/programs are available. Tools can be for example web-based, thereby providing for example a graphical HTML user interface to perform the analysis while the actual tool is running on a server. Standalone tools are tools you can download and install locally on your operating system or they can be launched directly from a web page through Java web Start (e.g. Artemis: Genome Browser and Annotation Tool, https://www.sanger.ac.uk/resources/software/artemis/). Data files with for example gene expression values can be opened with spreadsheet software such as Microsoft Excel or OpenOffice Calc (reference?). Programs and commands or scripts can also be run in Linux through a command-line interface.
The focus of this chapter will be the introduction to Linux as an operating system, the use of the command-line in Linux and commands.
An operating system (OS) is software developed to manage the processes and the interaction of the hardware itself and the hardware with the software. It also provides an environment in which programs can be run and developed. The operating system allows you to easily access your files and programs. Examples of such operating systems are Windows, Mac OS and Unix.
Unix is a multiuser operating system, which allows multitasking and networking. It uses a hierarchical file system and multiple small programs can interact through the shell. The shell is an interactive program in the operating system whereby a user can perform tasks (by typing commands with your keyboard) in a command-line interface and the shell communicates this to the operating system.
Through the command-line interface or terminal we can interact with the shell.
Linux is an open-source version of Unix, which is developed by Linus Torvalds (University of Helsinki). Several distributions of this open-source version exist, e.g. Ubuntu, Debian, Red Hat, SuSe, etc.
Both Unix and Linux use a command-line interface to execute tasks in the operating system. A command-line interface is provided by the shell and is used for executing command-line programs with commands and scripts. It also allows you to filter, sort and cut text files (e.g. GenBank format, XML format, etc.). Through the command-line interface programs can be run in batch, whereby multiple programs can run at the same time. It also allows distributed computing, which allows a process to be executed by multiple computers integrated in a network (e.g. LUDIT computing cluster, KU Leuven).
To be able to perform analysis on a Unix or Linux machine or a server, a connection needs to be made through the shell of the operating system. Through this shell we can use the command-line interface to execute tasks.
A server is a computer program that serves multiple computer programs and users. The computer(s) which run a server program are also called a server. It is often used for data storage and analysis processes, since it allows more computing power than performing analysis locally.
Here, we need to access the server machine through the shell of its operatins system, which is mostly Unix- and Linux-based. Access can be made with a Secure Shell (SSH) protocol, which allows communication with multiple computers in a network or a server. With Unix- and Linux-based computers (e.g. Mac OS) access can be immediately made with the terminal, which is a command-line interface that is provided by Unix and Linux. For computers working with the Windows operating system an intermediate software needs to be installed for accessing a Unix- or Linux-based computer/server. The software is called a Secure Shell Client (e.g. Bitvise SSH Client, https://www.bitvise.com/).
Host adress: bmw.gbiomed.kuleuven.be
Port: 22
Username: student-number, which is used for Toledo (r-number)

Bitvise SSH Client Login
A popup will show up to enter your password connected to your student-number (r-number).
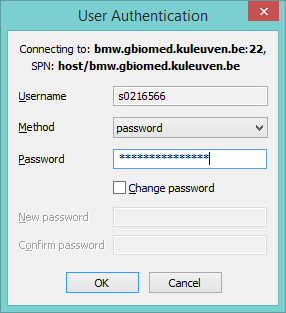
Bitvise SSH Client Login
When you are logged in two windows will pop up: Bitvise SFTP and Bitvise xterm.
With Bitvise SFTP you can view local files and remote files and transfer them from one to another by right clicking on the file and selecting 'Upload' or 'Download'. Thereby data, that is locally downloaded from a database, can be transferred from your computer to the remote file space in your own folder (/home/r-number) on the server to perform analysis. Afterwards, output data can be transferred back to your local file folder.

Bitvise Local and Remote folders
Bitvise xterm is your command-line interface, comparable with the terminal in Mac OS or Unix- and Linux-based systems. The command-line interface exists of a command prompt, which is a sequence of characters to inform the user that the command-line is able to accept commands. The command prompt mostly exists of the location of the user on the server and ends with a special character such as $.
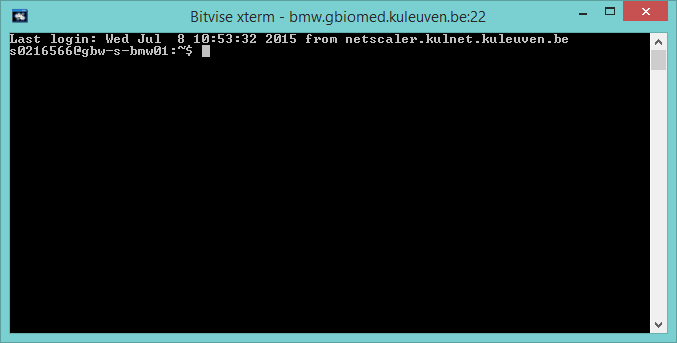
Bitvise SSH Client Command Line
Here, the command prompt exists of the username (s0123456) and the server adress (gbw-s-bmw01). In this sequence of characters the path to the current working directory is also given.
Files can be accessed and stored from different folders. Folders are made up of a hierarchical structure, which is comparable with graphical interfaces of e.g. Windows Explorer and
Folders are called directories and there are different levels:
Changing directories through the command-line can be done by typing cd 'change directory' in the command-line followed by the directory path which can be simple or more complex, e.g. cd /Bioinformatics or cd /Bioinformatics/Gene_expression/p53_data
Typing cd without a directory path takes you to the parent directory of your current working directory.
Two different types of pathnames exist. Absolute paths, which start from the root and displays the names of all the directories. Relative paths start from the working directory and use dots for representing directories. One dot (.) stands for the working directory and two dots (..) for the parent directory.

Absolute Relative Path
Hints and tricks:
The working directory can also be found in the command prompt. The full path to the current directory can be found by typing pwd in the command-line.
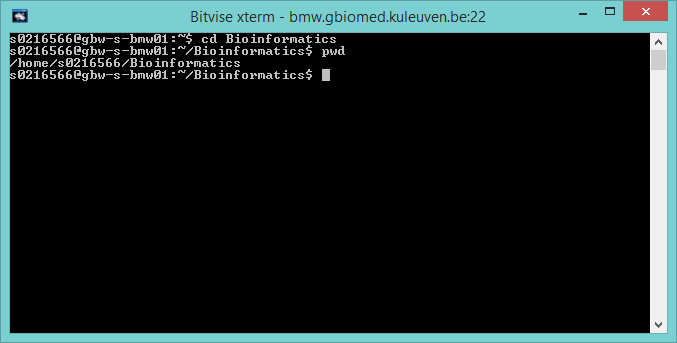
Bitvise SSH Client Command Line Folder
To have an overview of the files in your directory, simply type ls. View the characteristics, such as permissions, of all the files by ls -l. By specifying a path, you can view the content of distant directories.
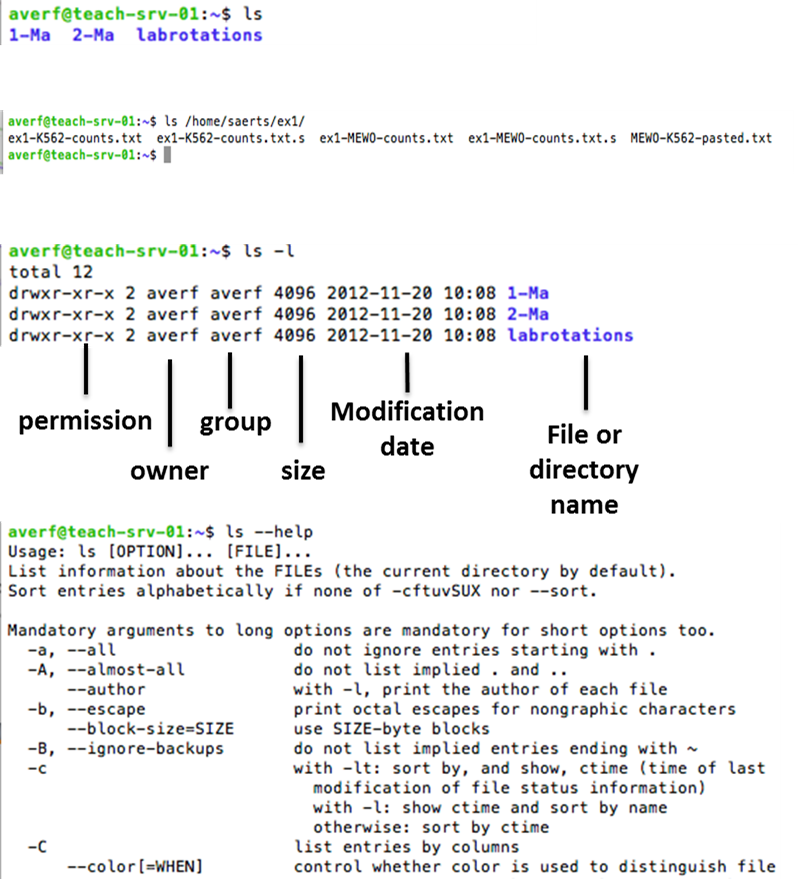
LS command
Following commands can also be used to handle files or directories:
g*: everything that starts with a g
*.txt: all file names with file extension .txt
mkdir: make a new directory, e.g. mkdir Gene_expression
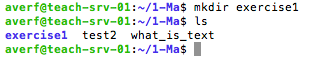
mkdir command
cp: copy file(s), use cp --h for options
Copy file1 into file2. If file2 does not exist, it is created. If it does exist it is overwritten:
cp file1 file 2If file2 exists, the option -i will ask the user to overwrite the file or not:
cp -i file1 file2Copy file1 into directory1:
cp file1 dir1Copy contents of directory1 into directory2 as its subdirectory named dir1. If dir2 does not exist, it is created:
cp -R dir1 dir2rm: remove file(s) permanently
mv: rename file(s), e.g. rename file called cell.txt with gene.txt; mv cell.txt gene.txt
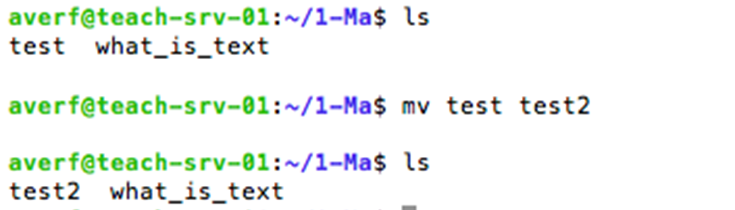
mv command
file: determine file type

file command
To view the file content less can be used.
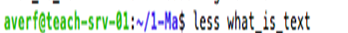
less command
Less can be navigated through the following:
Page Up or b: Scroll back one page
Page Down or space: Scroll forward one page
↓: Scroll down one line
h: Display help screen
q: Quit less
The directories/folders and files can also be viewed in the Bitvise SFTP window, where files can be transferred.
The general outline of commands in Linux is
command -options argumentsHints and tricks:
man before the command or --h after the command to get help and options with the command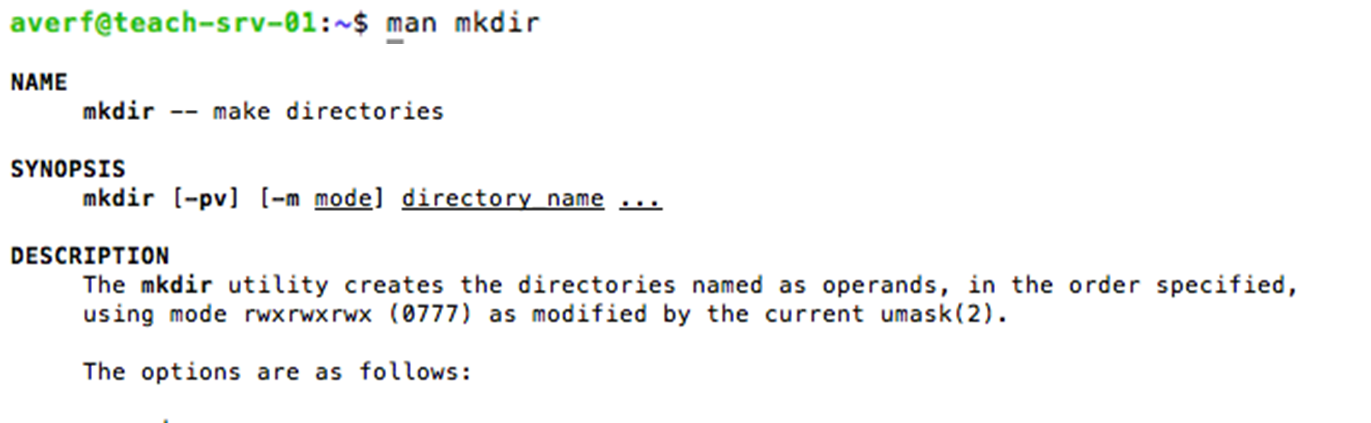
manual mkdir
Ctrl + c: interrupt a processCtrl + a: move the cursor to the beginning of the lineCtrl + e: move the cursor to the end of the lineCtrl + r: search for recently used commandsCtrl + l or type clear to clear the screen# in front of a command to not execute itLaunching a seperate screen from the command-line allows you to run commands which will not be terminated in its process when the screen and command-line is closed.
screen: open a screen (remote workspace)ctrl + a + d: detach the screenexit: terminate the screenscreen -r call up the screens that are running (with number)nano: an empy screen will appear
nano filename: file will open in a screen
Ctrl + o (^O): save file
Ctrl + x (^X): close nano
emacs filename: empty screen or your file will appear
Ctrl + x Ctrl + c: save and quit emacs
Redirecting input/output
Use > filename to put your results in an output file
Use >> filename to add results to an output file
Use < filename to use file as input
Use cat to concatenate files, it reads one or more files and copies them to a standard output
Use head and tail to show the first and last part of a file, respectively

head command
sort to sort lines of text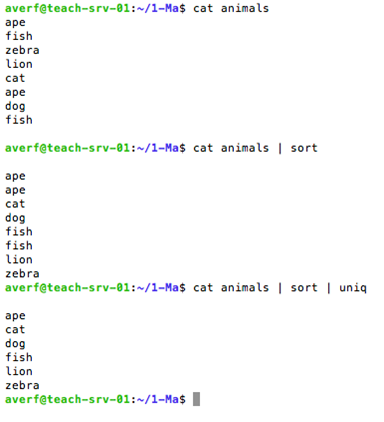
sort command
Use uniq to report or omit repeated lines
Use grep to print lines that match a pattern

grep command
wc to print line, word and bytes count for (each) file (wc --h)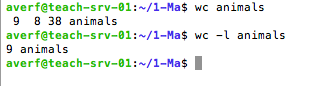
wc command
9: stands for the number of lines
8: stands for the number of words
38: stands for the number of bytes
locate to find a file by name
locate command
paste to merge lines of files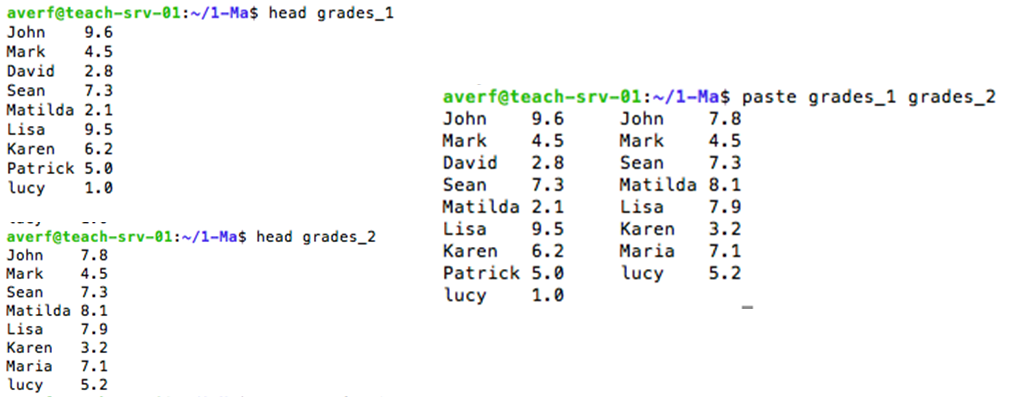
paste command
cut to remove/select sections from a file, cut -c: for the characters, cut -f: for the fields, cut -d: use delimiter as field seperator, cut --complement: everything except what is specified by -c and -f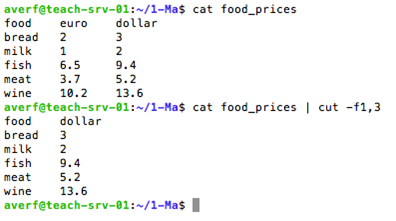
cut command
join to join the lines two files on a common field
join command
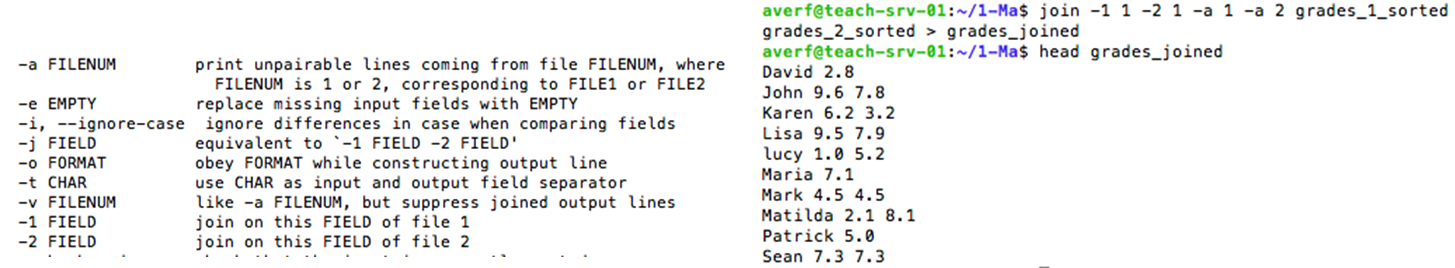
join manual
tr to translate or delete characters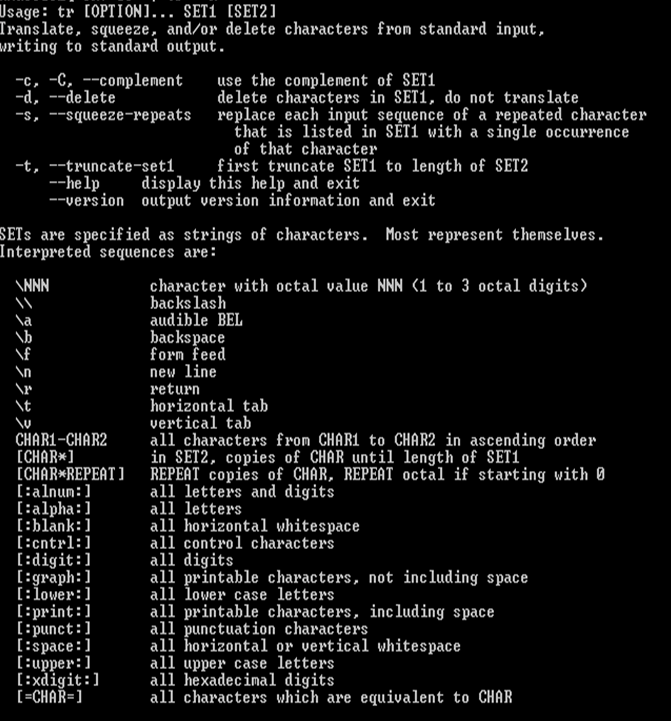
tr manual
tr command
comm to compare two sorted files line by line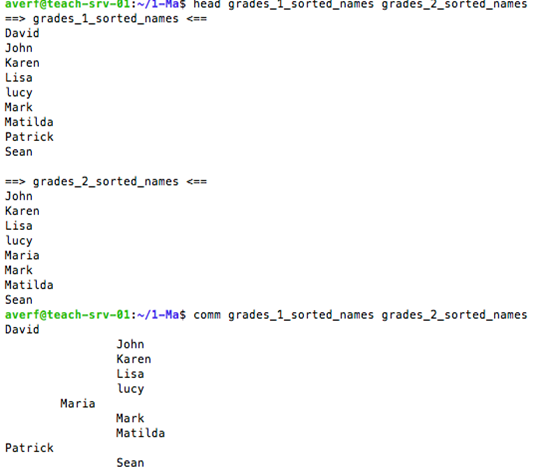
comm command
diff to compare files line by line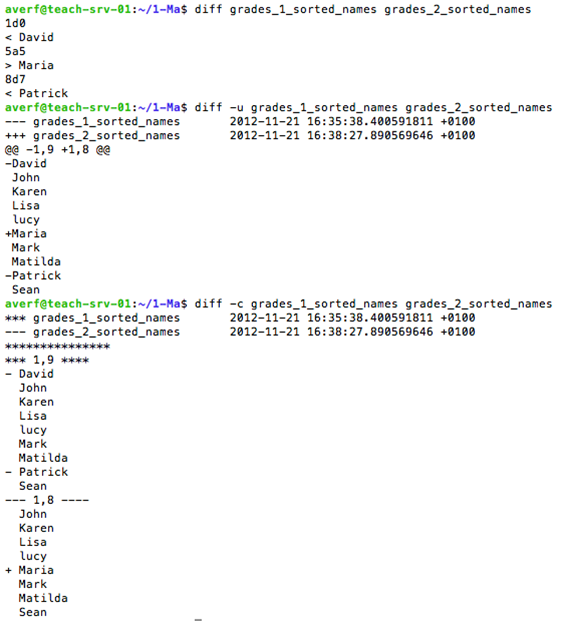
diff command
Hints and tricks:
| to perform a series of commands, with the output of the previous command used as the input for the following command| head after your command, before you execute it to check the format of the outputAdditional information:
Exercise 2.2. The use of commands for biological data interpretation
To exercise using commands in Linux at home, you can either install Linux (Ubuntu) and EMBOSS on your computer.
sudo apt-get install embossVia the KU Leuven network (Eduroam) you can login to the server. From your home network, just simply login to the server as you do at the KU Leuven network.
EMBOSS (The European Molecular Biology Open Software Suite) is a software analysis package (http://emboss.sourceforge.net/). Within EMBOSS many different applications are found for analysis of biological data such as sequence alignment, protein motif identification, nucleotide sequence pattern analysis, etc.
EMBOSS can be accessed in three ways:
EMBOSS
Galaxy