BioMart homepage
When performing gene expression analysis through microarrays or RNA-Seq, a list of expressed genes with their expression values is obtained as output data. One can analyze the functional enrichment of these large gene sets.
Performing gene expression analysis over multiple timepoints will give a list of genes which are expressed (higher or lower than control). Let's take a dataset of expression profiling of the differentiation of primary human skeletal myoblasts into myotubes during a time course (Warner JB, Philippakis AA, Jaeger SA, He FS, Lin J and Bulyk ML. Systematic identification of mammalian regulatory motifs' target genes and functions.Nat Methods. 2008;5(4):347-353.) Often, with expression data, clusters are generated to identify groups of genes with similar expression patterns.
Here, use cluster 0 (C0) from the gene expression dataset and obtain the identifier for each gene in this cluster(Supplementary Table 1).
Hints and tricks:
BioMart provides tools and serves as a platform to share and browse data (http://www.biomart.org/ or http://www.ensembl.org/biomart/martview/153013a85be70cb86b629f8ffa0e477c).

BioMart homepage
or
BioMart homepage
Here, we will convert the GenBank and RefSeq IDs from the dataset to UniProt IDs via BioMart on the Ensembl website.
In Filters, paste IDs and select the input format and output format (via Attributes -> External):
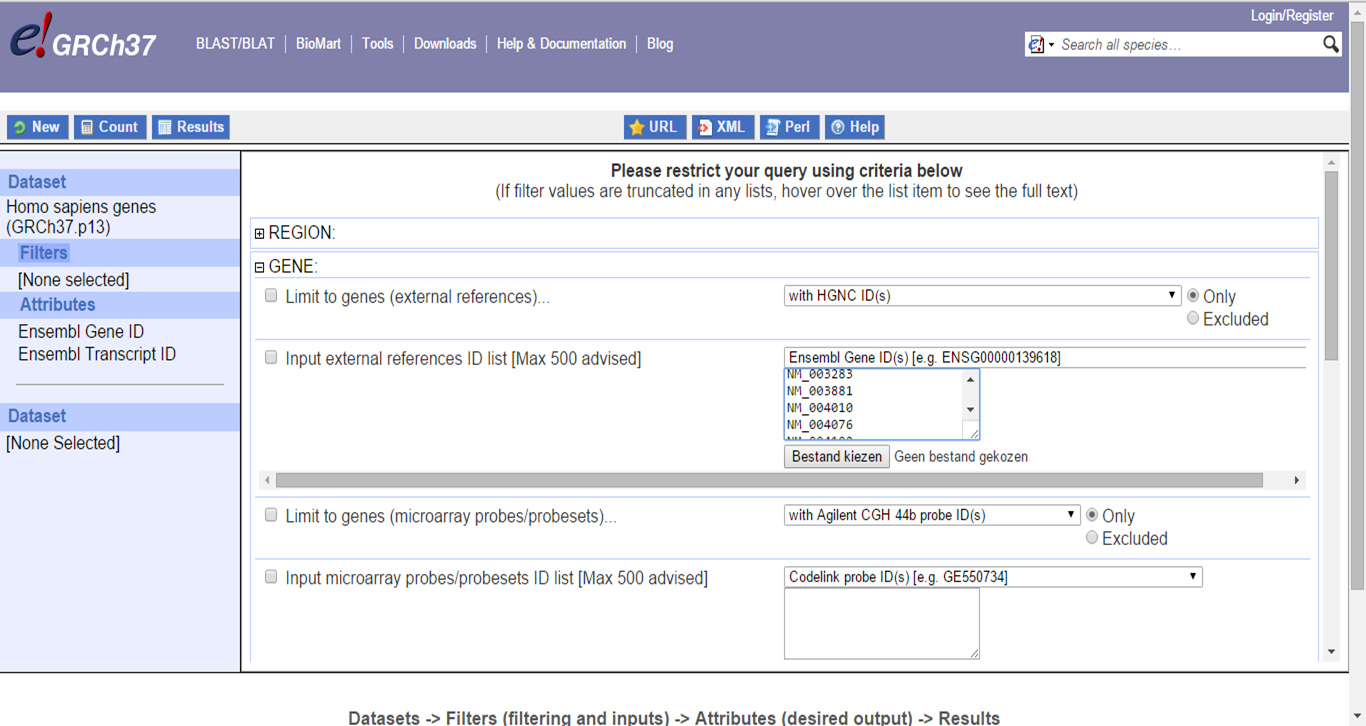
Gene IDs conversion

Gene IDs conversion
Click Results on top to see the converted IDs:
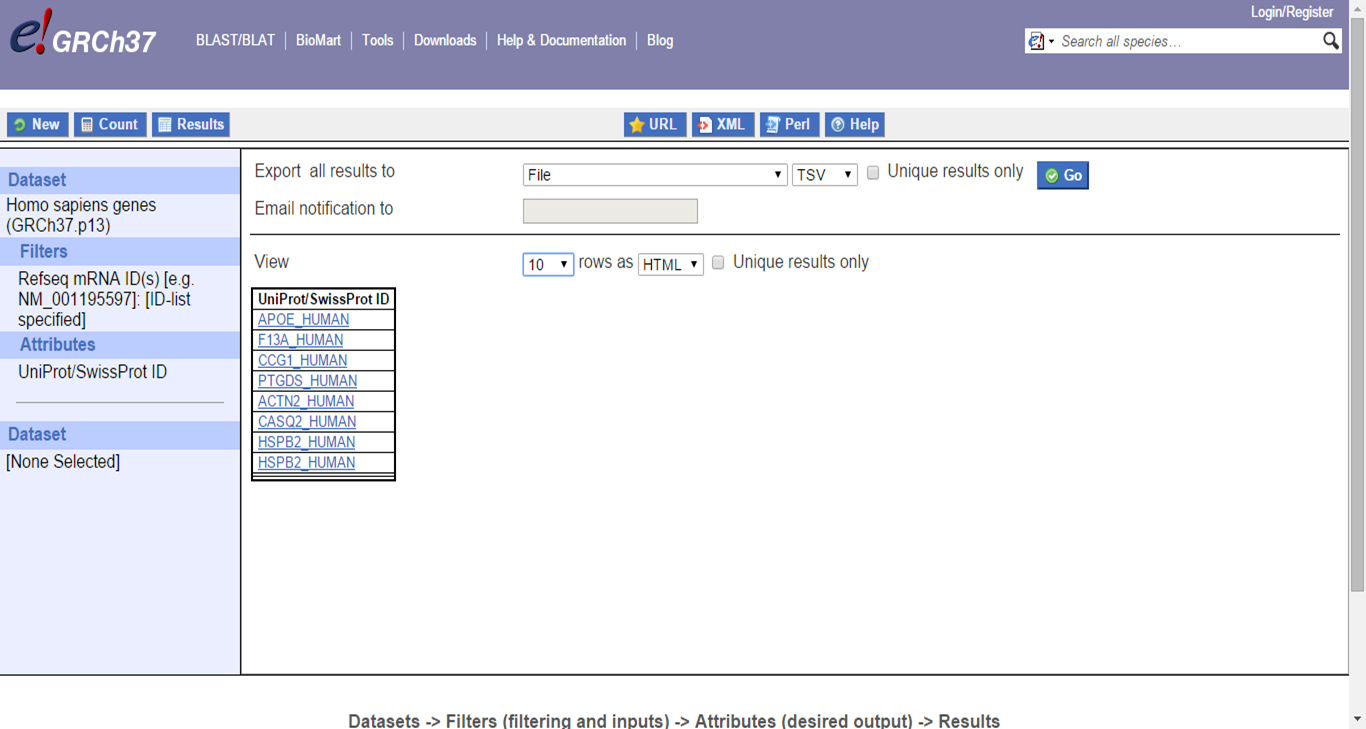
Gene list IDs conversion results
BioMart.org offers an ID conversion tool:
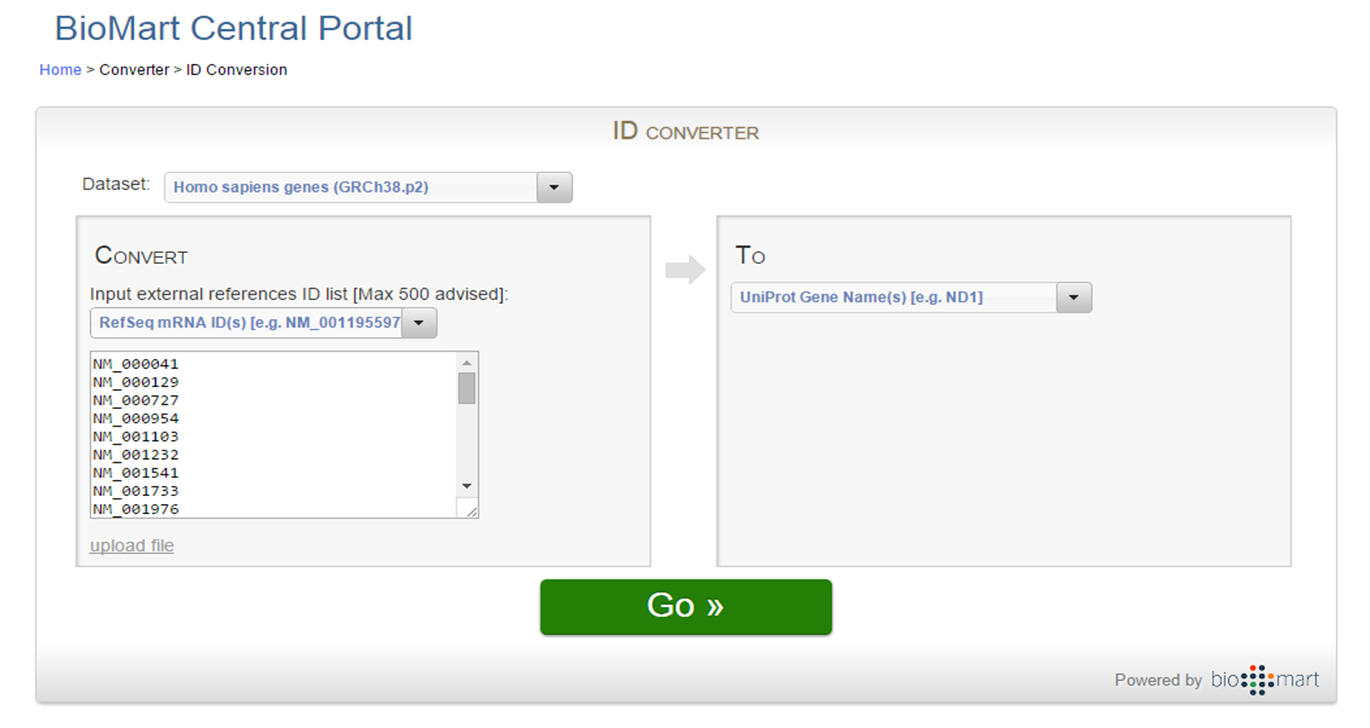
BioMart ID Conversion
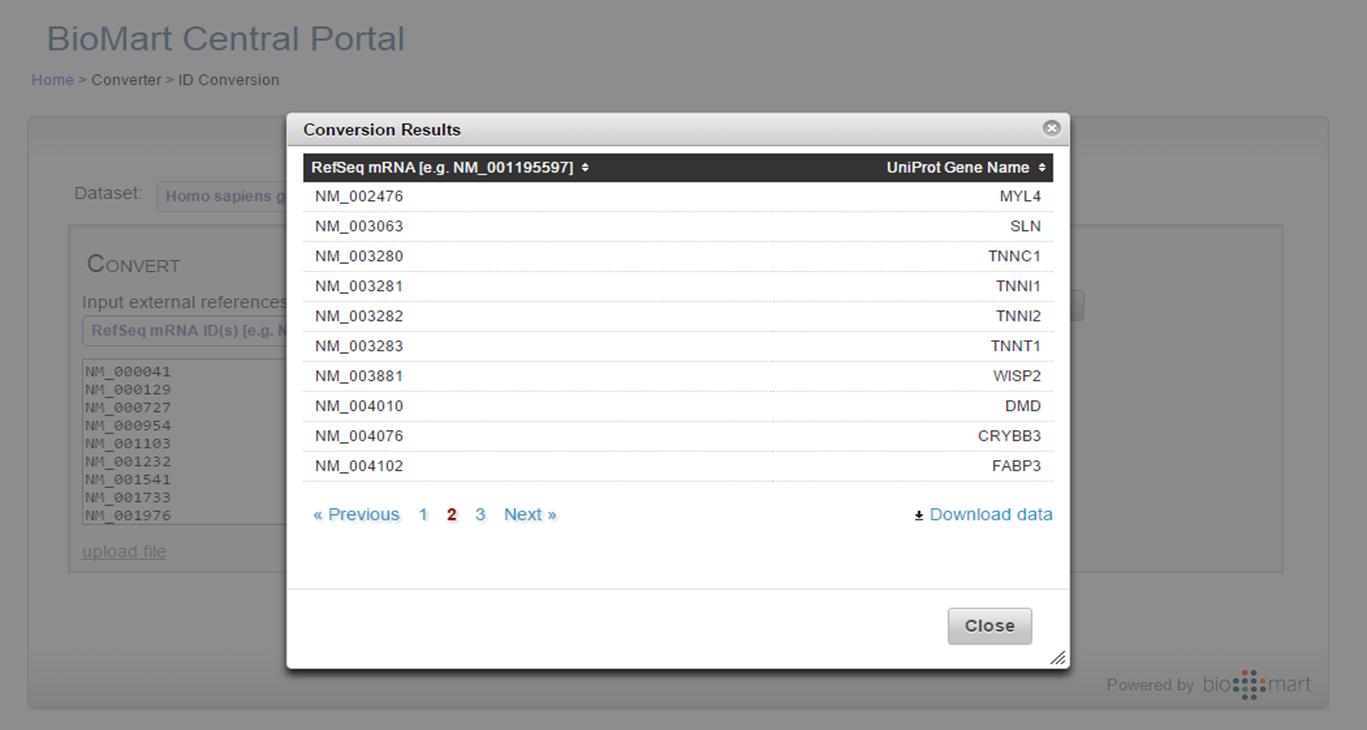
BioMart ID Conversion output
BioMart also allows for identification of orthologs for different species:
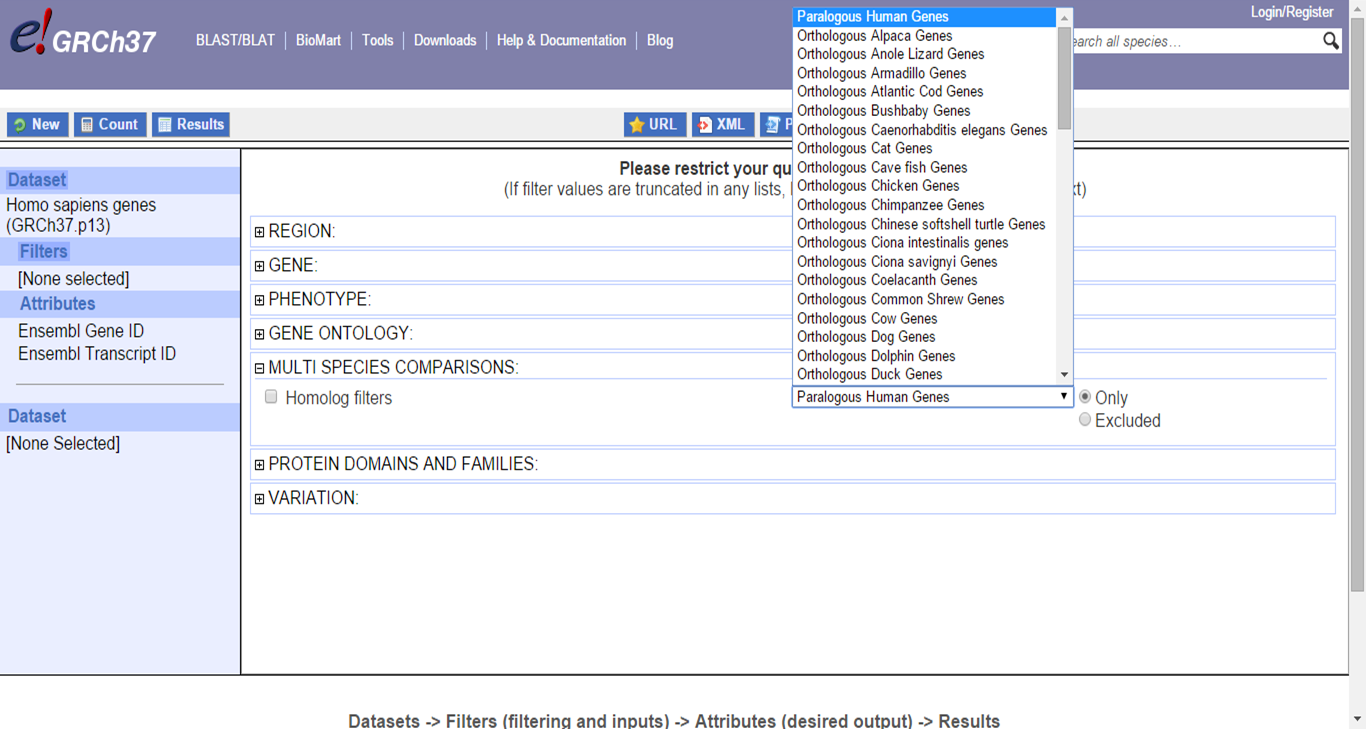
BioMart Homologous genes
You can also download certain sequence attributes from specific genes, for example 3'UTR:
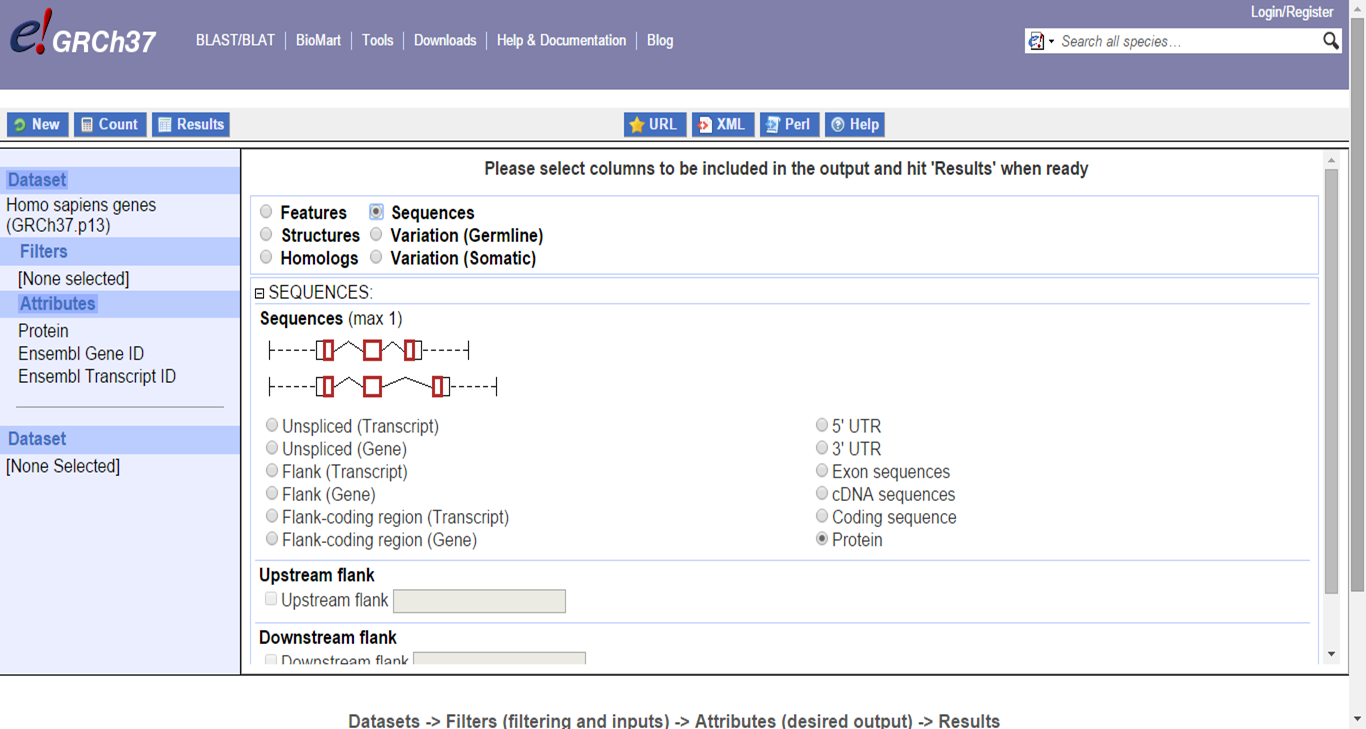
BioMart sequence retrieval
Now, we have obtained the UniProt IDs we can perform Gene Ontology Enrichment / functional enrichment analysis. Therefore, we can use Gene Ontology (http://geneontology.org/) or Webgestalt (http://bioinfo.vanderbilt.edu/webgestalt/).
Gene Ontology enrichment analysis
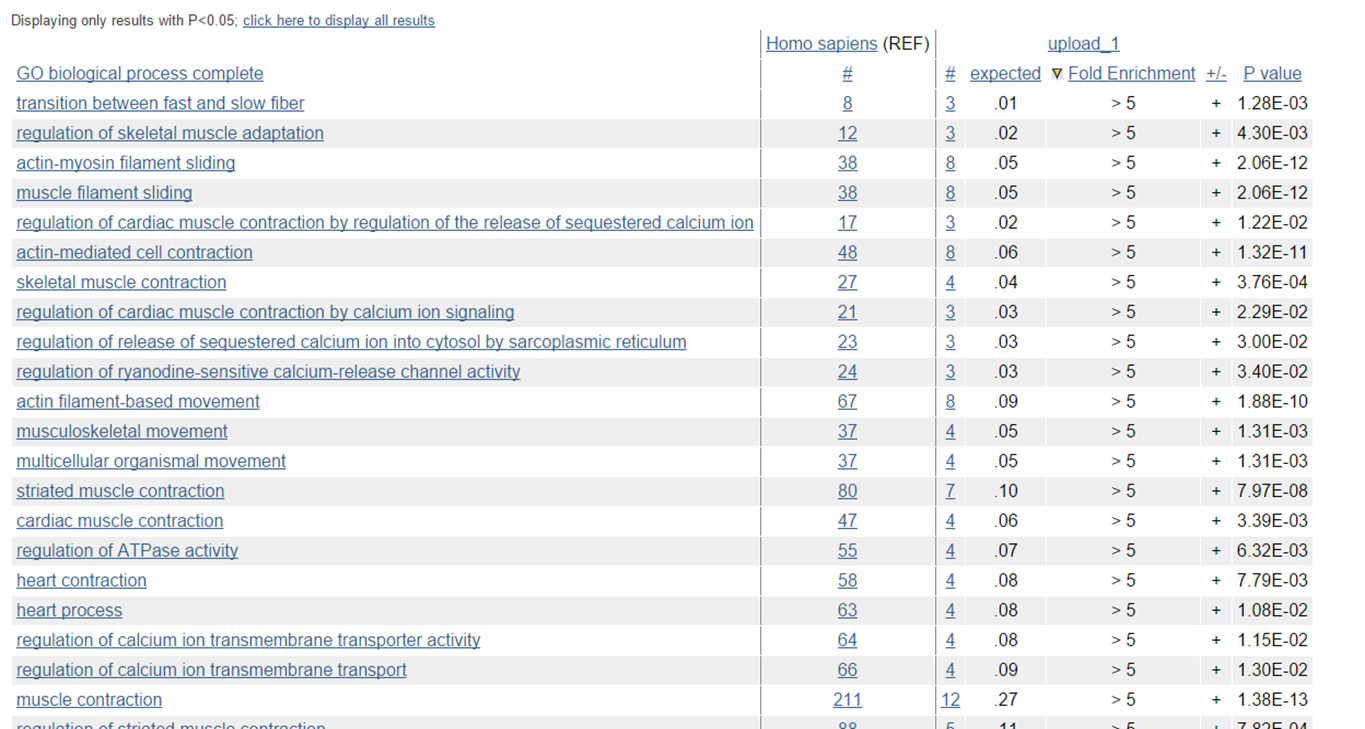
GO output